Manuscripts
The manuscripts listed here are those affiliated with CompBioClub. For complete lists, see the members' scholar pages.
- BPformer: An Interpretable Deep Learning Framework for Livestock Breed Proportion AnalysisJinpeng Wang, Shuo Sun, Yaran Zhang, Zhihua Ju, Qiang Jiang, Xiuge Wang, Yao Xiao, Lingxi Chen*, and Jin Ming Huang*Preprint-In Submission, 2025
Introduction: Breed proportion analysis plays a crucial role in cattle genetic resource conservation and breeding improvement. With the rapid development of genomic technologies, breed proportion prediction based on single nucleotide polymorphisms (SNPs) has become a current research hotspot. However, existing methods still face challenges such as insufficient interpretability and the urgent need for feature engineering. Methods: This study developed the BPformer model, which combines convolutional neural networks and self-attention mechanisms, specifically designed for livestock breed proportion prediction. We utilized SNP data from 15 Chinese indigenous cattle breeds and 12 foreign commercial breeds, employing 39,868 high-quality SNPs loci as the gold standard dataset. Dimensionality-reduced datasets were constructed through four feature selection methods (FST, In, BP_AVE, and BP_GRA). The study compared the performance of BPformer against traditional machine learning models (SVR, KNR, and RF) and other deep learning models (MLP, and CNN) on the dimensionality-reduced datasets, while performance evaluation of the three deep learning models was conducted on the gold standard dataset. Results: BPformer outperformed other models across all four detection methods with BBP SNPs = 4,000 and in the gold standard testing scenarios. Through attention mechanism visualization and SHAP value analysis, we identified key SNPs loci that contributed most significantly to the prediction of each breed proportion component, thereby enhancing the model’s interpretability. Conclusion: BPformer effectively addresses the interpretability challenges faced by traditional methods from a modeling perspective and can efficiently capture long-range dependencies among SNPs loci. This provides a powerful tool for Chinese cattle breed resource conservation and genomic selection breeding, which is of great significance for maintaining genetic diversity in Chinese livestock industry.
@article{BPformer, title = {BPformer: An Interpretable Deep Learning Framework for Livestock Breed Proportion Analysis}, author = {Wang, Jinpeng and Sun, Shuo and Zhang, Yaran and Ju, Zhihua and Jiang, Qiang and Wang, Xiuge and Xiao, Yao and Chen, Lingxi and Huang, Jin Ming}, journal = {Preprint-In Submission}, year = {2025}, doi = {10.21203/rs.3.rs-8340493/v1}, peerreviewed = {false} } -
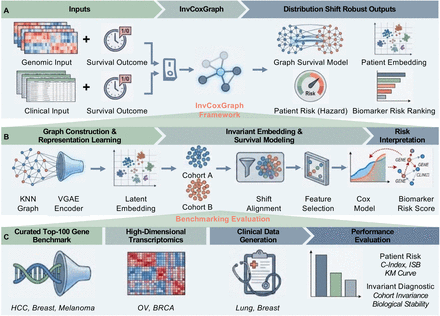 Learning Invariant Graph Representations for Cox Survival Modeling under Distribution ShiftsKa Ho Ng†, Chengshang Lyu†, Anna Jiang, Yinhu Li, and Lingxi Chen*Preprint-In Submission, 2025
Learning Invariant Graph Representations for Cox Survival Modeling under Distribution ShiftsKa Ho Ng†, Chengshang Lyu†, Anna Jiang, Yinhu Li, and Lingxi Chen*Preprint-In Submission, 2025Survival prediction from high-dimensional biomedical data is frequently compromised by distribution shifts across multi-center cohorts, where models trained on specific populations often rely on spurious correlations that fail to generalize to new environments. While recent independence-driven reweighting techniques attempt to mitigate this, they typically treat patients as isolated instances, neglecting the intrinsic topological structures and biological pathways shared within patient populations. To address this limitation, we propose InvGraphCox (Invariant Graph Cox), a novel framework that integrates graph-structured representation learning with robust survival modeling. InvGraphCox constructs a k-nearest-neighbor patient graph to capture local manifold structures and employs a Variational Graph Autoencoder (VGAE) combined with a cohort-wise alignment mechanism to learn low-dimensional patient embeddings that are invariant to site-specific biases. We comprehensively evaluate the framework across three distinct experimental settings: the Curated Top-100 Gene Benchmark for stable biomarker identification, large-scale, high-dimensional transcriptomic datasets (Ovarian and Breast Cancer) for unsupervised representation learning, and clinical datasets (Breast and Lung Cancer) involving mixed-type covariates. Experimental results demonstrate that InvGraphCox consistently outperforms state-of-the-art baselines in terms of discrimination, calibration, and risk stratification, confirming its ability to extract robust, biologically meaningful representations in heterogeneous healthcare settings.
@article{InvCoxGraph, title = {Learning Invariant Graph Representations for Cox Survival Modeling under Distribution Shifts}, author = {Ng, Ka Ho and Lyu, Chengshang and Jiang, Anna and Li, Yinhu and Chen, Lingxi}, journal = {Preprint-In Submission}, year = {2025}, doi = {10.64898/2025.11.30.691365}, peerreviewed = {false} } -
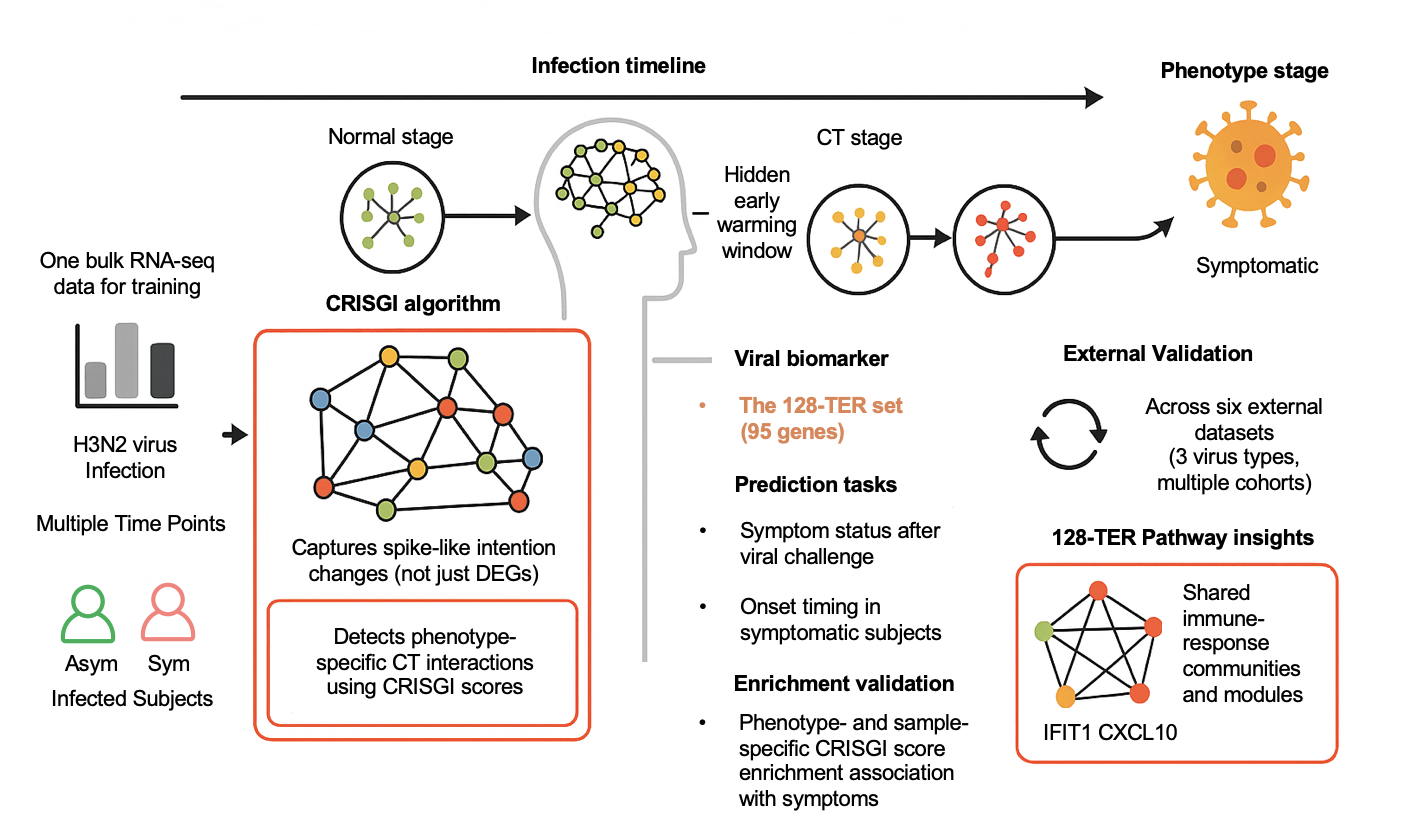 Predicting Early Transitions in Respiratory Virus Infections via Critical Transient Gene InteractionsChengshang Lyu, Anna Jiang, Ka Ho Ng, Xiaoyu Liu, and Lingxi Chen*Preprint-Under Review, 2025
Predicting Early Transitions in Respiratory Virus Infections via Critical Transient Gene InteractionsChengshang Lyu, Anna Jiang, Ka Ho Ng, Xiaoyu Liu, and Lingxi Chen*Preprint-Under Review, 2025Early detection of respiratory virus infections, such as influenza A (H3N2), is critical for timely intervention and disease management. Conventional biomarkers often overlook the complex and dynamic nature of gene regulatory changes, while existing predictive models frequently lack automation and robust external validation. Thus, we present CRISGI (Critical tran-Sient Gene Interaction), a computational framework that detects early-warning signals of infection by identifying dynamic changes in gene-gene interactions—termed critical transient interactions—from bulk RNA-seq data. CRISGI leverages critical transition (CT) theory to capture a GRN’s unstable intermediate state, known as the CT stage, before irreversible phenotypic shifts. Applied to a human challenge study with H3N2, CRISGI identified 128 critical transition edges (128-TER). These were used to train predictive models capable of forecasting symptom status and onset timing. 128-TER was then validated across six temporal transcriptomic datasets involving three respiratory viruses (H3N2, H1N1, HRV). The 128-TER consistently distinguished symptomatic individuals, predicted infection onset, and revealed phenotype-specific enrichment patterns. Notably, CRISGI captured immune-related transitions involving interferon-stimulated genes (e.g., IFIT1, CXCL10), underscoring their role in early host defense. CRISGI advances early-warning biomarker discovery by integrating interaction-level dynamics and predictive modeling. Its reproducibility across viruses highlights shared immune activation pathways, supporting its utility in both research and clinical contexts.
@article{CRISGI, title = {Predicting Early Transitions in Respiratory Virus Infections via Critical Transient Gene Interactions}, author = {Lyu, Chengshang and Jiang, Anna and Ng, Ka Ho and Liu, Xiaoyu and Chen, Lingxi}, journal = {Preprint-Under Review}, year = {2025}, doi = {10.1101/2025.04.18.649619}, peerreviewed = {false}, } -
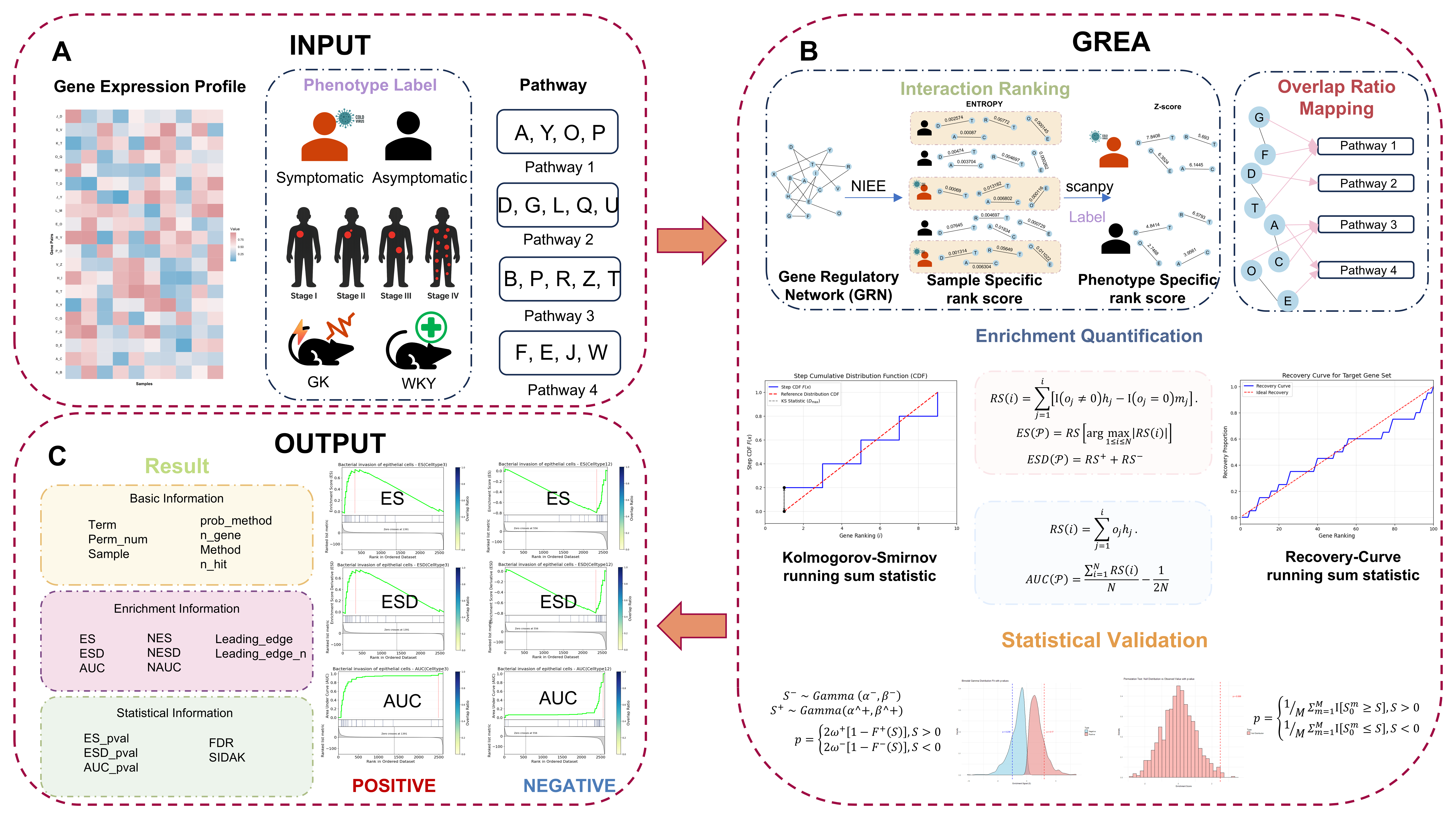 Knowledge-driven annotation for gene interaction enrichment analysisXiaoyu Liu†, Anna Jiang†, Chengshang Lyu, and Lingxi Chen*Preprint-Under Review, 2025
Knowledge-driven annotation for gene interaction enrichment analysisXiaoyu Liu†, Anna Jiang†, Chengshang Lyu, and Lingxi Chen*Preprint-Under Review, 2025Gene Set Enrichment Analysis (GSEA) is a cornerstone for interpreting gene expression data, yet traditional approaches overlook gene interactions by focusing solely on individual genes, limiting their ability to detect subtle or complex pathway signals. To overcome this, we present GREA (Gene Interaction Enrichment Analysis), a novel framework that incorporates gene interaction data into enrichment analysis. GREA replaces the binary gene hit indicator with an interaction overlap ratio, capturing the degree of overlap between gene sets and gene interactions to enhance sensitivity and biological interpretability. It supports three enrichment metrics: Enrichment Score (ES), Enrichment Score Difference (ESD) from a Kolmogorov-Smirnov-based statistic, and Area Under the Curve (AUC) from a recovery curve. GREA evaluates statistical significance using both permutation testing and gamma distribution modeling. Benchmarking on transcriptomic datasets related to respiratory viral infections shows that GREA consistently outperforms existing tools such as blitzGSEA and GSEApy, identifying more relevant pathways with greater stability and reproducibility. By integrating gene interactions into pathway analysis, GREA offers a powerful and flexible tool for uncovering biologically meaningful insights in complex datasets. The source code is available at https://github.com/compbioclub/GREA.
@article{GREA, title = {Knowledge-driven annotation for gene interaction enrichment analysis}, author = {Liu, Xiaoyu and Jiang, Anna and Lyu, Chengshang and Chen, Lingxi}, journal = {Preprint-Under Review}, year = {2025}, doi = {10.1101/2025.04.15.649030}, peerreviewed = {false}, } -
 Biologically Informative NA Deconvolution (BIND) excavates hidden features of the proteome from missing values in large-scale datasetsWeiheng Guo†, Wenyi Jin†, Jieyi Zheng†, Yilin Pan, Rui Wang, Jian Zhang*, Xikang Feng*, Lingxi Chen*, and Liang Zhang*Preprint-Under Revision, 2025
Biologically Informative NA Deconvolution (BIND) excavates hidden features of the proteome from missing values in large-scale datasetsWeiheng Guo†, Wenyi Jin†, Jieyi Zheng†, Yilin Pan, Rui Wang, Jian Zhang*, Xikang Feng*, Lingxi Chen*, and Liang Zhang*Preprint-Under Revision, 2025The fast-advancing mass spectrometry and related technologies have greatly extended the depth of coverage in large-scale proteomics studies, including single-cell applications. As sample numbers grow rapidly, it is often challenging to interpret the proteins with missing values that are often presented as “NA” (not available). It could be the evidence of no expression, low expression below the detection threshold, or false negative detection due to technical issues. Existing methods for missing values imputation, while generally useful, rarely consider the non-random NA values that inform biological significance. In the current study, we developed Biologically Informative NA Deconvolution (BIND) that applies an adaptive neighborhood-based modeling to deconvolve the nature of NAs as “biological” (low/no expression) or technical (experimental errors). Applying to multiple cell line datasets and human tissue extracellular vesicle datasets, BIND excavated the NAs that indicated “hallmark absence” of unique proteins. This led to improvements in protein-protein interaction analysis and the identification of novel disease biomarkers. To facilitate its public accessibility, we compiled BIND into a web server that features functional online operations and interactive visualizations. Furthermore, we demonstrated that the BIND server could deconvolve the NAs and improve the analyses of single-cell proteomics datasets. Overall, BIND delineates the biological significance of missing values rather than treating them as a burden, providing a critical perspective for understanding the complex proteome in various biological contexts.
@article{BIND, title = {Biologically Informative NA Deconvolution (BIND) excavates hidden features of the proteome from missing values in large-scale datasets}, author = {Guo, Weiheng and Jin, Wenyi and Zheng, Jieyi and Pan, Yilin and Wang, Rui and Zhang, Jian and Feng, Xikang and Chen, Lingxi and Zhang, Liang}, journal = {Preprint-Under Revision}, year = {2025}, doi = {10.1101/2025.06.19.660508}, peerreviewed = {false} }